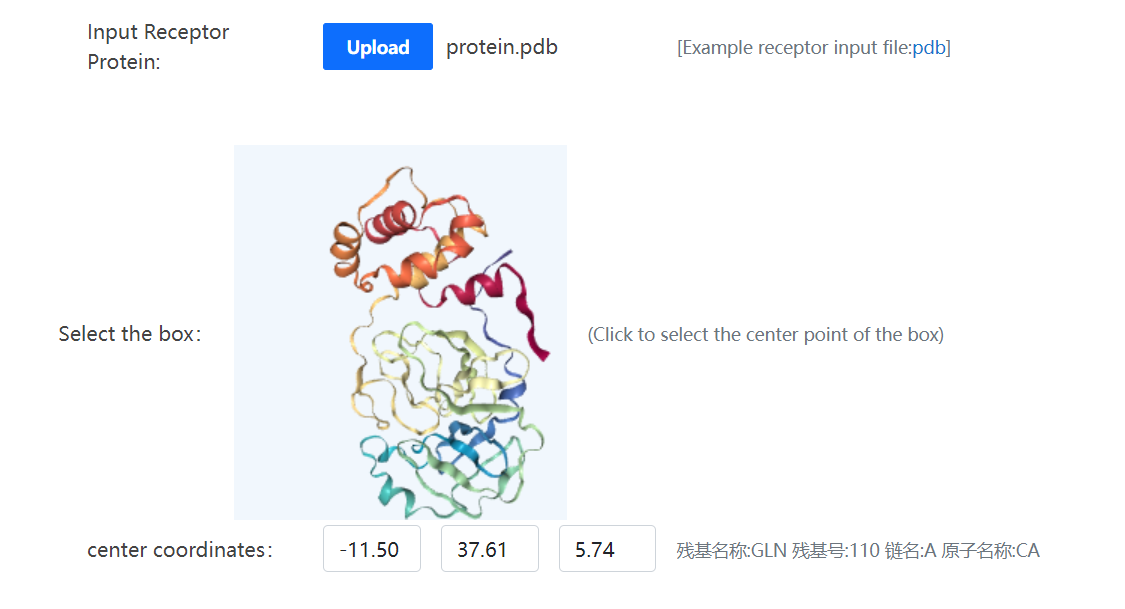
You need to select a receptor protein target and upload it in a strict pdb format. After the upload is complete, the receptor protein will be displayed as NGL(License: NGL)attempt, you need to click on the protein select box center point. The center coordinates you selected are displayed below, along with the residue name, residue number, chain name, and atom name.
Next you need to select a target and upload small molecules in strict mol2 and sdf formats. Open Babel (License: Open Babel Official Page) is used for format transformation or 3D coordinate generation for the uploaded files. Tempdock is used as docking engine. An email will be sent to your email address with a direct link to access your docking results.
After your job completes, you could enter the job ID to access the result page.
Alternatively, you can access the docking results by the corresponding link in your email. The download url is provided as: http://codockligand.schanglab.org.cn/index/result/index.php?job_id=*************
When the job is completed, the server will send an email to the user. By default, the server only displays the top 10 models. The receptor protein target is renamed rec.pdb, the ligand protein target is renamed raw_1.mol2. The top 10 models are named 0.sdf-9.sdf, respectively. The models can be visualized in 3D by NGL (License: NGL). The scored file is named as scores.txt. You can view and download your docking results from the result page.
If you have any suggestions or questions, please Contact Us.